Implementation of Nanotechnology with DNA
- Sarbojit Das
- Aug 11, 2021
- 3 min read
Introduction
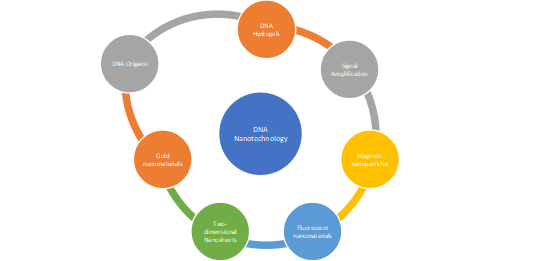
Nanotechnology is the purist’s approach to biomolecular engineering. This field aims to create molecular structures and devices through the exclusive use of DNA as an engineering material1. The well-characterized nature of DNA base pairing provides an easy means to control DNA interactions. The success of DNA nanotechnology comes from three key ingredients:
our quantitative understanding of DNA thermodynamics, which makes it possible to predict how single-stranded DNA molecules fold and interact with one another,
the rapidly falling cost and increasing quality of DNA synthesis, and
the focus on cell-free settings, where designed reaction pathways can proceed without interference from DNA and RNA processing enzymes that might be encountered in cells.
DNA nanotechnology has long been motivated by the goal of building ‘smart therapeutics’, drug delivery systems, tools for molecular biology and other devices that could interact with or operate within living cells. Such applications play to the obvious strengths of nucleic acid nanostructures and devices, particularly their small size, biocompatibility and straightforward manner in which they could be programmed to interact with cellular nucleic acids through hybridization.
Cell-free DNA nanotechnology
To operate reliably in complex, wet environments, living organisms use sensory receptors to detect changes in that environment, motors and actuators to adapt to the environment, computational control circuits to convert sensor information into motor activity, and structural elements that protect and organize these components. Intriguingly, cell-free DNA nanotechnology has made progress towards the construction of most of the functional components — both structures and dynamic devices — required for creating molecular ‘robots’ that can emulate some of the behavioural complexity observed in biology. [2]
DNA nanotechnology in lysates and fixed cells
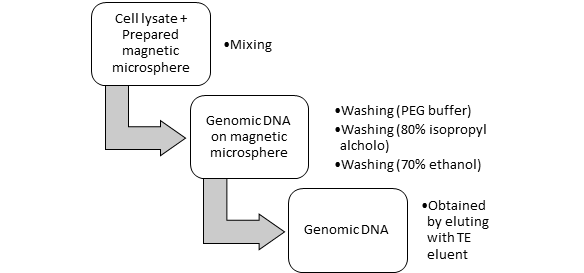
Cellular conditions are considerably different from that of cell-free experiments. The presence of nucleic-acid-binding proteins, including DNases and RNases, may interfere with device performance, and cellular environments are highly structured, which inhibits the free diffusion of exogenously delivered nucleic acids. Cell lysates, serum and fixed cells provide reaction environments that each capture some of the complexity of live cells and enable testing and optimization of nucleic acid devices in well-controlled conditions.[3]
Conclusion
DNA-based therapeutics and diagnostics are set apart from more established approaches because of their capacity to respond to the surrounding environment. Molecular logic and conditional (un)hiding of drug moieties could decrease side effects and increase specificity. Even the relatively simple one- or two-input systems built so far have resulted in increased specificity and performance, and could be further improved with more complex multi-input logic. Diagnostic and therapeutic decisions are routinely based on the analysis of panels of multiple molecular markers, be they proteins, RNA, DNA, lipids, sugars or metabolites. For example, immunologists must often consider large numbers of cell surface proteins to delineate all of the various cell types in a blood sample. Gene expression classifiers that reliably distinguish different tissues and disease states are typically built on measurements of tens or hundreds of different RNA species. Given the success of dynamic DNA nanotechnology in scaling up the size and reliability of molecular circuits in cell-free settings, it is intriguing to think that DNA ‘biocomputers’ could eventually perform complex diagnostic tasks based on the analysis of tens of molecular markers directly in living organisms.
References
Seeman, N. C. & Belcher, A. M. Emulating biology: building nanostructures from the bottom up. Proc. Natl Acad. Sci. USA 99, 6451–6455 (2002).
Chen, J. H. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991).
Fu, T. J. & Seeman, N. C. DNA double-crossover molecules. Biochemistry 32, 3211–3220 (1993).
By Sarbojit Das
sarbojit.25@gmail.com



Comments